Robert Hänsel-Hertsch
Epigenetic determinants of genome stability for mammalian tissue
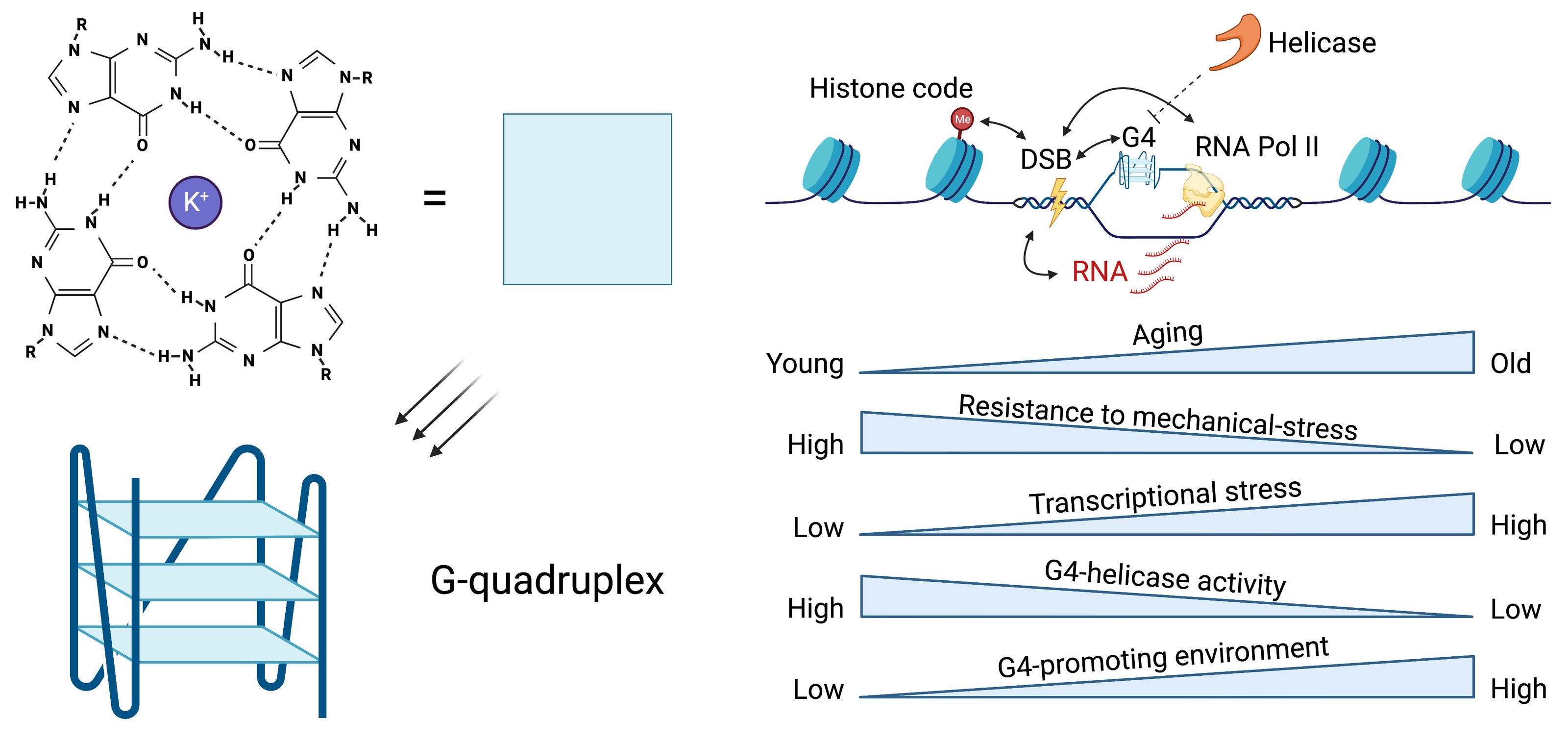
This project will elucidate epigenetic mechanisms promoting genome instability when tissue homeostasis declines with age and under physiological stress. We discovered elevated G-quadruplex (G4) DNA secondary structure formation in mutated highly transcribed gene regulatory regions of cancer, suggesting dysregulated G4 DNA regions as promoter of genome instability. We have and continue to gather evidence that dysregulated G4s emerge before cancer development in physiologically aged murine tissues and rapidly aged murine and human models. We hypothesize here that dysregulated G4 secondary structure formation may promote genome instability and heterostasis in aged and stress-induced tissues of rodent models. To address this and determine related mechanisms, we will employ our newly developed genome-wide multiomics technology to jointly map epigenetically linked DNA breakage beyond association. To identify epigenetic regulators that promote an increase in epigenetically linked DNA breakage in tissues of our rodent models, we will use computational predictions and experimental associations of our multiomics data with existing data sets. We will further use proximity ligation proteomics to directly link regulatory factors to epigenetically linked DNA breakage and validate these findings by genetic loss-of-function studies in C. elegans.
PROJECT RELATED PUBLICATIONS
- Hunold P*, Hoehne MN*, Kiljan M, van Ray O, Herter J, Herter-Sprie GS, Hänsel-Hertsch R. G-quadruplex DNA structures mediate non-autonomous instruction of breast tumour microenvironments. bioRxiv 2023 https://doi.org/10.1101/2023.01.16.524243. *contributed equally
- Hänsel-Hertsch R, Simeone A, Shea A, Hui WWI, Zyner KG, Marsico G, Rueda OM, Bruna A, Martin A, Zhang X, Adhikari S, Tannahill D, Caldas C, Balasubramanian S. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat Genet. 2020 Sep;52(9):878-883. doi: 10.1038/s41588-020-0672-8. Epub 2020 Aug 3. PMID: 32747825
- Hänsel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018 Mar;13(3):551-564. doi: 10.1038/nprot.2017.150. Epub 2018 Feb 22. PMID: 29470465
- Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol. 2017 May;18(5):279-284. doi: 10.1038/nrm.2017.3. Epub 2017 Feb 22. PMID: 28225080
- Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016 Oct;48(10):1267-72. doi: 10.1038/ng.3662. Epub 2016 Sep 12. PMID: 27618450

